statmechcrack.isotensional
A module for the crack model in the isotensional ensemble.
This module consist of the class CrackIsotensional which
contains methods for computing quantities in the
isotensional (constant force) thermodynamic ensemble.
- class CrackIsotensional(**kwargs)[source]
Bases:
CrackIsometricThe crack model class for the isotensional ensemble.
- Z_0_isotensional(p, lambda_)[source]
The nondimensional isotensional partition function as a function of the nondimensional end force for the reference system,
\[Z_0(p,\boldsymbol{\lambda}) = Z_b(p,\lambda_1,\lambda_2) e^{-\beta U_{01}(\boldsymbol{\lambda})}.\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The nondimensional isotensional partition function.
- Return type:
numpy.ndarray
Example
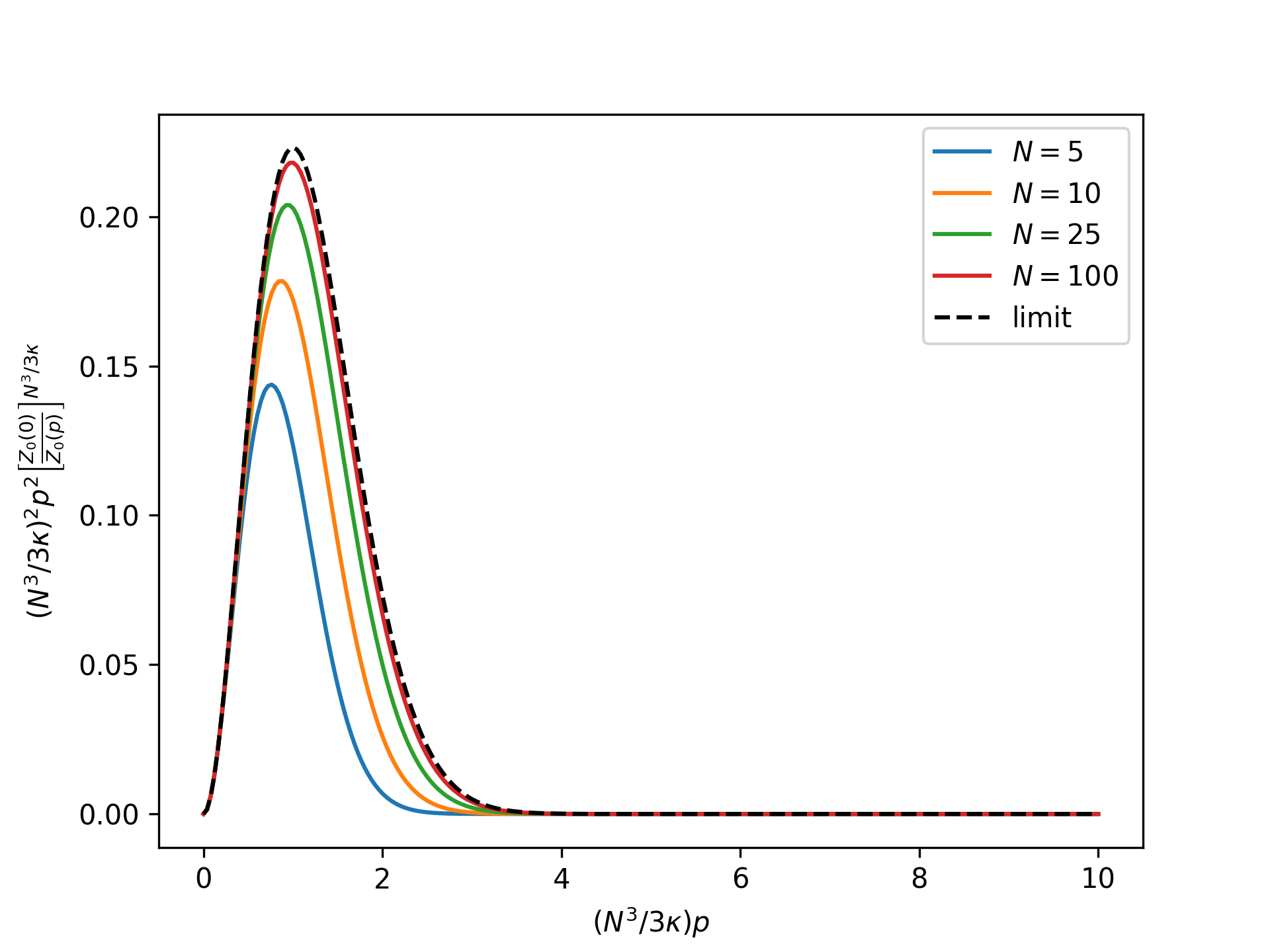
Plot the rescaled nondimensional equilibrium radial distribution as a function of the nondimensional end force for the reference system in the isotensional ensemble for an increasing number of broken bonds \(N\) and compare to the thermodynamic limit:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 250) >>> _ = plt.figure() >>> for N in [5, 10, 25, 100]: ... model = CrackIsotensional(N=N) ... p = 3*model.kappa/N**3*rp ... r_Z_0 = (model.Z_0_isotensional(0, [1, 1]) ... / model.Z_0_isotensional(p, [1, 1]) ... )**(model.N**3/3/model.kappa) ... _ = plt.plot(rp, rp**2*r_Z_0, label='$N=$'+str(N)) >>> _ = plt.plot(rp, rp**2*np.exp(-rp*(rp/2 + 1)), ... 'k--', label='limit') >>> _ = plt.xlabel(r'$(N^3/3\kappa)p$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)^2p^2$' + ... r'$\left[\frac{Z_0(0)}{Z_0(p)}\right]^{N^3/3\kappa}$') >>> _ = plt.legend() >>> plt.show()
- Z_b_isotensional(p, lambda_)[source]
The nondimensional isotensional partition function as a function of the nondimensional end separation for the isolated bending system,
\[Z_b(p,\lambda_1,\lambda_2) = \sqrt{\frac{(2\pi)^{N+1}}{\det\mathbf{H}}} \,e^{\frac{1}{2}\mathbf{g}^T\cdot\mathbf{H}^{-1}\cdot\mathbf{g}-f},\]where \(\mathbf{H}\), the nondimensional Hessian of the total potential energy for the isolated bending system, and \(\mathbf{g}\) and \(f\) are
\[\mathbf{g}(p,\lambda_1,\lambda_2) = \kappa\left( p/\kappa, 0, \ldots, 0, -\lambda_1, 4\lambda_1 - \lambda_2 \right)^T,\]\[f(\lambda_1,\lambda_2) = \frac{\kappa}{2}\left[\lambda_1^2 + \left(2\lambda_1 - \lambda_2\right)^2\right] .\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The nondimensional isotensional partition function.
- Return type:
numpy.ndarray
- Z_isotensional(p, transition_state=False)[source]
The nondimensional isotensional partition function as a function of the nondimensional end force,
\[Z(p) = \int d\lambda \ Z_0(p,\boldsymbol{\lambda}) e^{-\beta U_1(\boldsymbol{\lambda})},\]approximated using the asymptotic relation
\[Z(p) \sim Z_0(p, \hat{\boldsymbol{\lambda}}) \prod_{j=1}^M \sqrt{\frac{2\pi}{\beta u''(\hat{\lambda}_j)}} \, e^{-\beta u(\hat{\lambda}_j)},\]which is valid for \(\varepsilon\gg 1\), where \(\hat{\boldsymbol{\lambda}}\) is from minimizing \(\beta\Pi\).
- Parameters:
p (array_like) – The nondimensional end force.
approach (str, optional, default='asymptotic') – The calculation approach.
transition_state (bool, optional, default=False) – Whether or not to calculate in the transition state.
Example
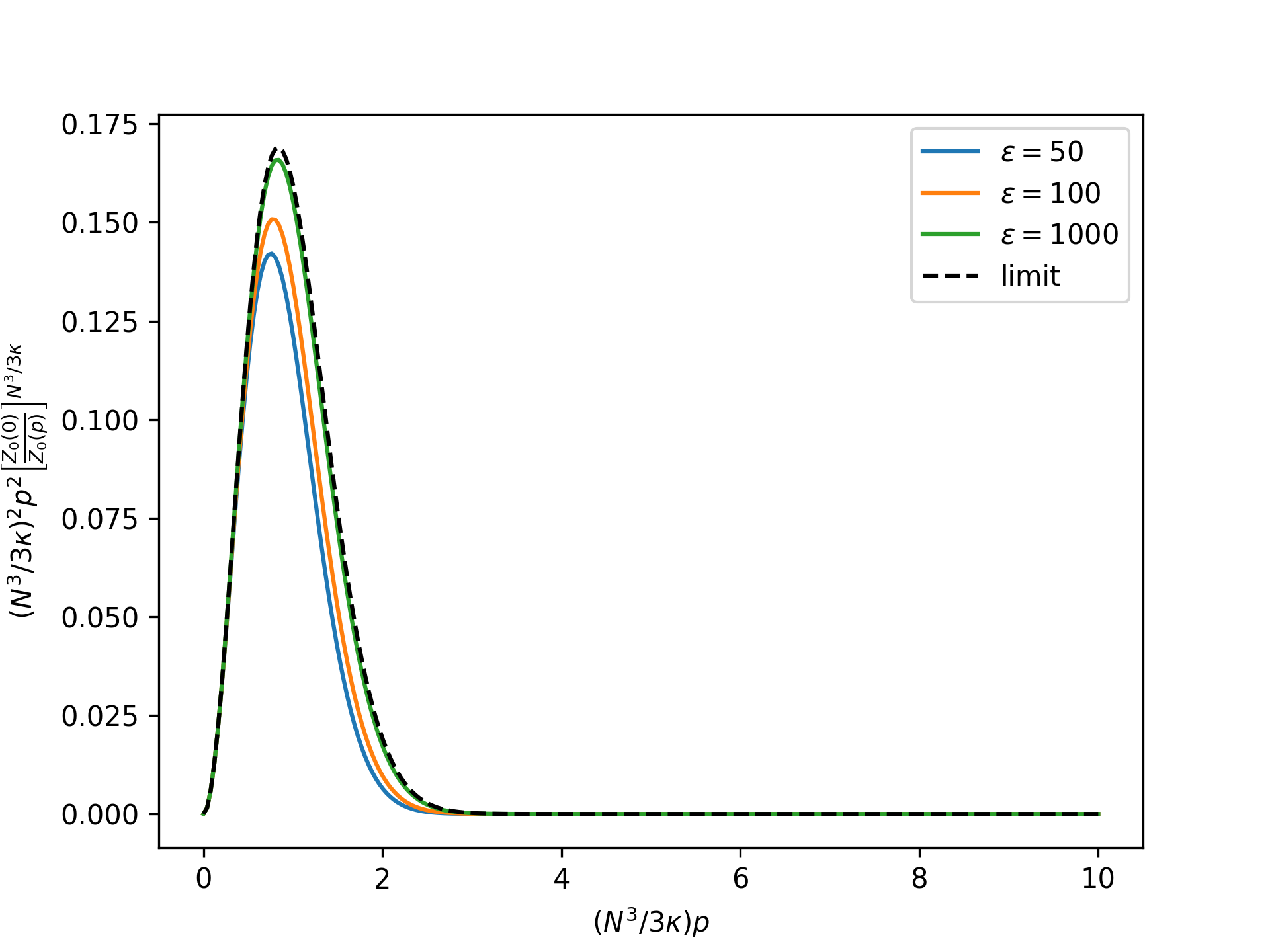
Plot the rescaled nondimensional equilibrium radial distribution as a function of the nondimensional end force in the isotensional ensemble for an increasing nondimensional bond energy and compare to the limit given by the reference system:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 250) >>> _ = plt.figure() >>> for varepsilon in [50, 100, 1000]: ... model = CrackIsotensional(varepsilon=varepsilon) ... p = 3*model.kappa/model.N**3*rp ... r_Z = (model.Z_isotensional(0)/model.Z_isotensional(p) ... )**(model.N**3/3/model.kappa) ... _ = plt.plot(rp, rp**2*r_Z, ... label=r'$\varepsilon=$'+str(varepsilon)) >>> r_Z_0 = (model.Z_0_isotensional(0, [1, 1]) ... / model.Z_0_isotensional(p, [1, 1]) ... )**(model.N**3/3/model.kappa) >>> _ = plt.plot(rp, rp**2*r_Z_0, 'k--', label='limit') >>> _ = plt.xlabel(r'$(N^3/3\kappa)p$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)^2p^2$' + ... r'$\left[\frac{Z_0(0)}{Z_0(p)}\right]^{N^3/3\kappa}$') >>> _ = plt.legend() >>> plt.show()
- Returns:
The nondimensional isotensional partition function.
- Return type:
numpy.ndarray
- beta_G_0_abs_isotensional(p, lambda_)[source]
The absolute nondimensional Gibbs free energy as a function of the nondimensional end force for the reference system,
\[\beta G_0(p,\boldsymbol{\lambda}) = -\ln Z_0(p,\boldsymbol{\lambda}) = \beta G_b(p,\lambda_1,\lambda_2) + \beta U_{01}(\boldsymbol{\lambda}).\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The absolute nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
- beta_G_0_isotensional(p, lambda_)[source]
The relative nondimensional Gibbs free energy as a function of the nondimensional end force for the reference system,
\[\beta\Delta G_0(p,\boldsymbol{\lambda}) = \beta G_0(p,\boldsymbol{\lambda}) - \beta G_0(0,\boldsymbol{\lambda}).\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The relative nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
Example
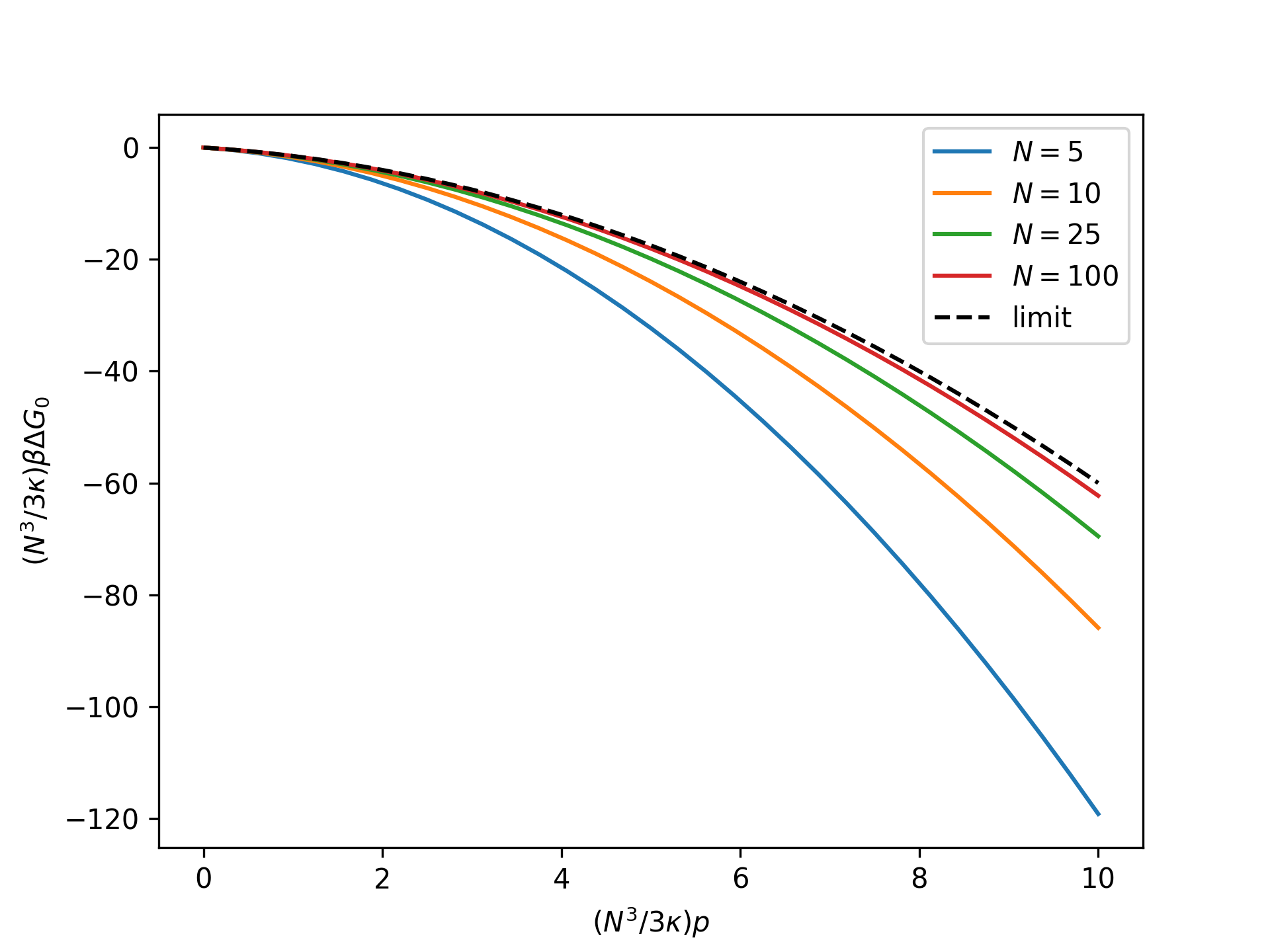
Plot the rescaled nondimensional relative Gibbs free energy as a function of the rescaled nondimensional end force for the reference system in the isotensional ensemble for an increasing number of broken bonds \(N\) and compare to the thermodynamic limit:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 33) >>> _ = plt.figure() >>> for N in [5, 10, 25, 100]: ... model = CrackIsotensional(N=N) ... p = 3*model.kappa/N**3*rp ... beta_G_0 = \ ... model.beta_G_0_isotensional(p, [1, 1]) ... _ = plt.plot(rp, N**3/3/model.kappa*beta_G_0, ... label='$N=$'+str(N)) >>> _ = plt.plot(rp, -rp*(rp/2 + 1), 'k--', label='limit') >>> _ = plt.xlabel(r'$(N^3/3\kappa)p$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)\beta\Delta G_0$') >>> _ = plt.legend() >>> plt.show()
- beta_G_abs_isotensional(p, approach='asymptotic', transition_state=False)[source]
The absolute nondimensional Gibbs free energy as a function of the nondimensional end force,
\[\beta G(p) = -\ln Z(p).\]- Parameters:
p (array_like) – The nondimensional end force.
approach (str, optional, default='asymptotic') – The calculation approach.
transition_state (bool, optional, default=False) – Whether or not to calculate in the transition state.
- Returns:
The absolute nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
- beta_G_b_abs_isotensional(p, lambda_)[source]
The absolute nondimensional Gibbs free energy as a function of the nondimensional end force for the isolated bending system,
\[\beta G_b(p,\lambda_1,\lambda_2) = -\ln Z_b(p,\lambda_1,\lambda_2).\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The absolute nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
- beta_G_b_isotensional(p, lambda_)[source]
The relative nondimensional Gibbs free energy as a function of the nondimensional end force for the isolated bending system,
\[\beta\Delta G_b(p,\lambda_1,\lambda_2) = \beta G_b(p,\lambda_1,\lambda_2) - \beta G_b(0,\lambda_1,\lambda_2).\]In the thermodynamic limit of a large number of broken bonds \(N\), this function has the asymptotic relation
\[\beta\Delta G_b(p,\lambda_1,\lambda_2) \sim -\frac{N^3}{6\kappa}\,p^2 - p \quad\text{for }N\gg 1.\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The relative nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
- beta_G_isotensional(p, approach='asymptotic', **kwargs)[source]
The relative nondimensional Gibbs free energy as a function of the nondimensional end force,
\[\beta\Delta G(p) = \beta G(p) - \beta G(0).\]- Parameters:
p (array_like) – The nondimensional end force.
approach (str, optional, default='asymptotic') – The calculation approach.
**kwargs – Arbitrary keyword arguments. Passed to
beta_G_isotensional_monte_carlo().
- Returns:
The relative nondimensional Gibbs free energy.
- Return type:
numpy.ndarray
Example
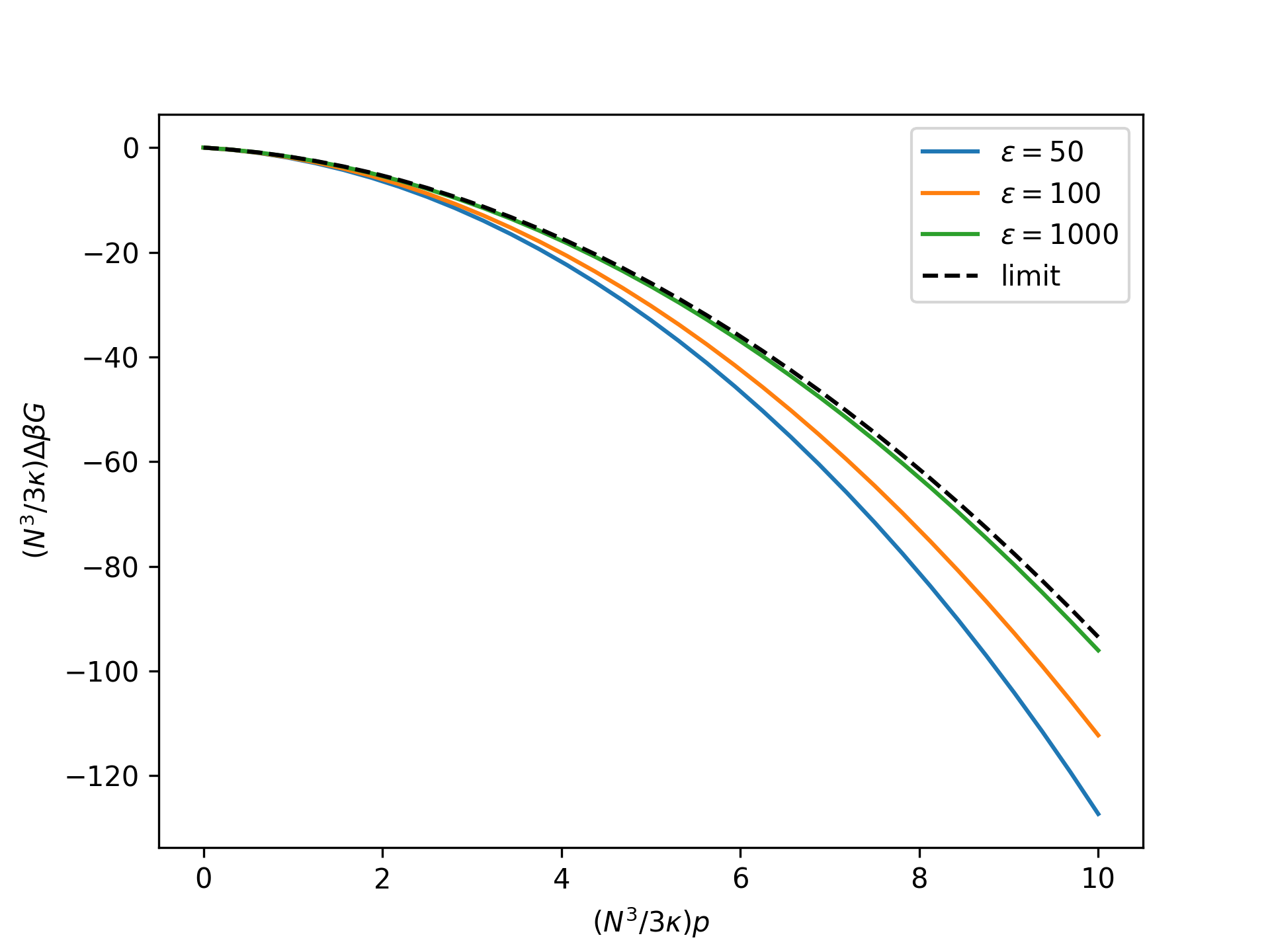
Plot the rescaled nondimensional relative Gibbs free energy as a function of the rescaled nondimensional end force in the isotensional ensemble for an increasing nondimensional bond energy and compare to the limit given by the reference system:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 33) >>> _ = plt.figure() >>> for varepsilon in [50, 100, 1000]: ... model = CrackIsotensional(varepsilon=varepsilon) ... p = 3*model.kappa/model.N**3*rp ... beta_G = model.beta_G_isotensional( ... p, approach='asymptotic') ... _ = plt.plot(rp, model.N**3/3/model.kappa*beta_G, ... label=r'$\varepsilon=$'+str(varepsilon)) >>> beta_G_0 = model.beta_G_0_isotensional(p, [1, 1]) >>> _ = plt.plot(rp, model.N**3/3/model.kappa*beta_G_0, ... 'k--', label='limit') >>> _ = plt.xlabel(r'$(N^3/3\kappa)p$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)\Delta\beta G$') >>> _ = plt.legend() >>> plt.show()
- k_0_isotensional(p, lambda_)[source]
The nondimensional forward reaction rate coefficient as a function of the nondimensional end force for the reference system in the isotensional ensemble.
- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The nondimensional forward reaction rate.
- Return type:
numpy.ndarray
- k_b_isotensional(p, lambda_)[source]
The nondimensional forward reaction rate coefficient as a function of the nondimensional end force for the isolated bending system in the isotensional ensemble.
- Parameters:
p (array_like) – The nondimensional end force.
lambda (array_like) – The intact bond stretches.
- Returns:
The nondimensional forward reaction rate.
- Return type:
numpy.ndarray
- k_isotensional(p, approach='asymptotic', **kwargs)[source]
The nondimensional forward reaction rate coefficient as a function of the nondimensional end force in the isotensional ensemble.
- Parameters:
p (array_like) – The nondimensional end force.
approach (str, optional, default='asymptotic') – The calculation approach.
- Returns:
The nondimensional forward reaction rate.
- Return type:
numpy.ndarray
Example
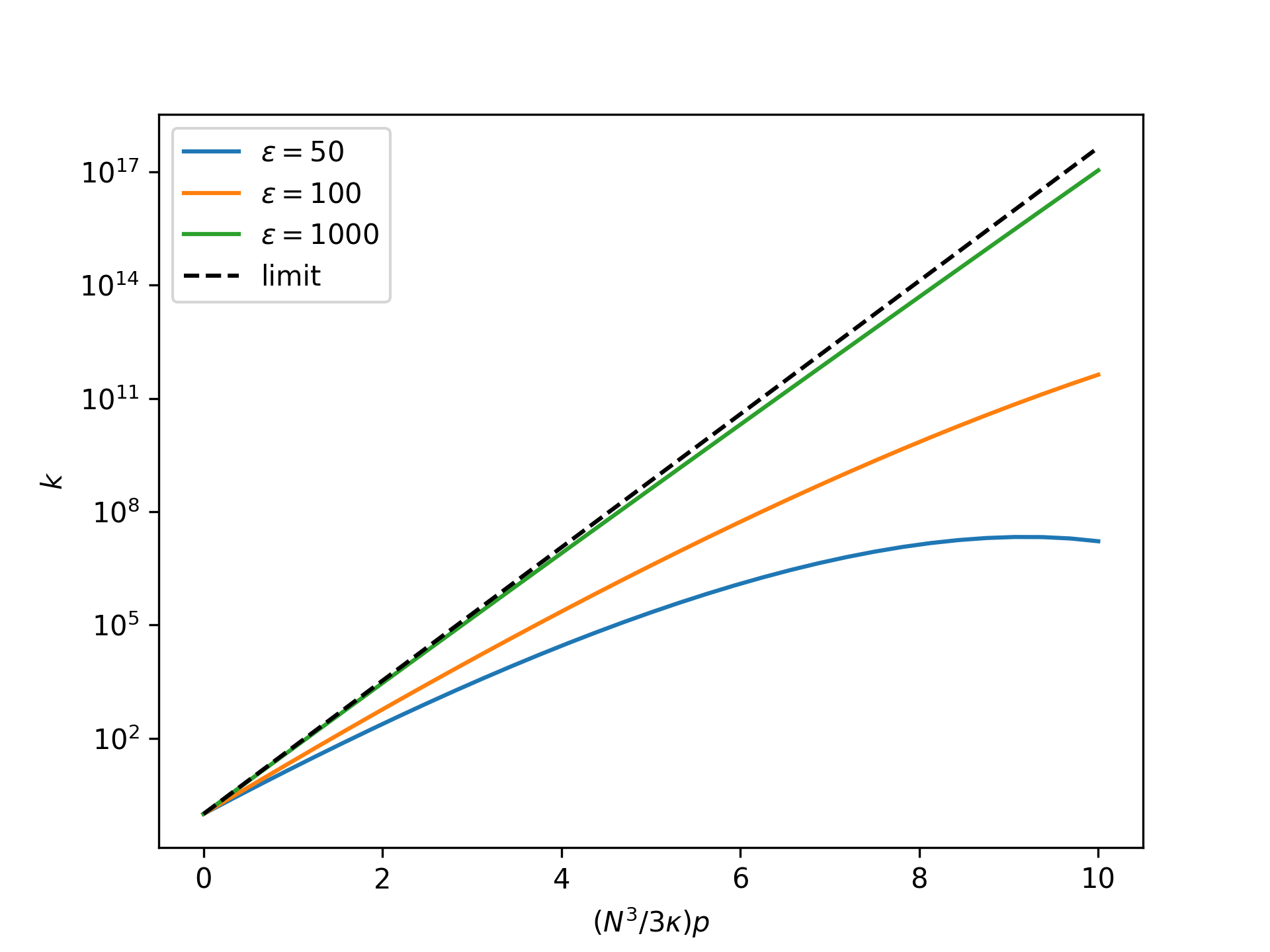
Plot the nondimensional forward reaction rate coefficient as a function of the nondimensional end force in the isotensional ensemble for an increasing nondimensional bond energy and compare to the limit given by the reference system:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 33) >>> _ = plt.figure() >>> for varepsilon in [50, 100, 1000]: ... model = CrackIsotensional(varepsilon=varepsilon) ... p = 3*model.kappa/model.N**3*rp ... _ = plt.semilogy( ... rp, model.k_isotensional(p), ... label=r'$\varepsilon=$'+str(varepsilon)) >>> _ = plt.semilogy(rp, model.k_0_isotensional(p, [1, 1]), ... 'k--', label='limit') >>> _ = plt.xlabel(r'$(N^3/3\kappa)p$') >>> _ = plt.ylabel(r'$k$') >>> _ = plt.legend() >>> plt.show()
- k_net_isotensional(p)[source]
The nondimensional net reaction rate coefficient as a function of the nondimensional end force in the isotensional ensemble.
- Parameters:
p (array_like) – The nondimensional end force.
- Returns:
The nondimensional net reaction rate.
- Return type:
numpy.ndarray
Example
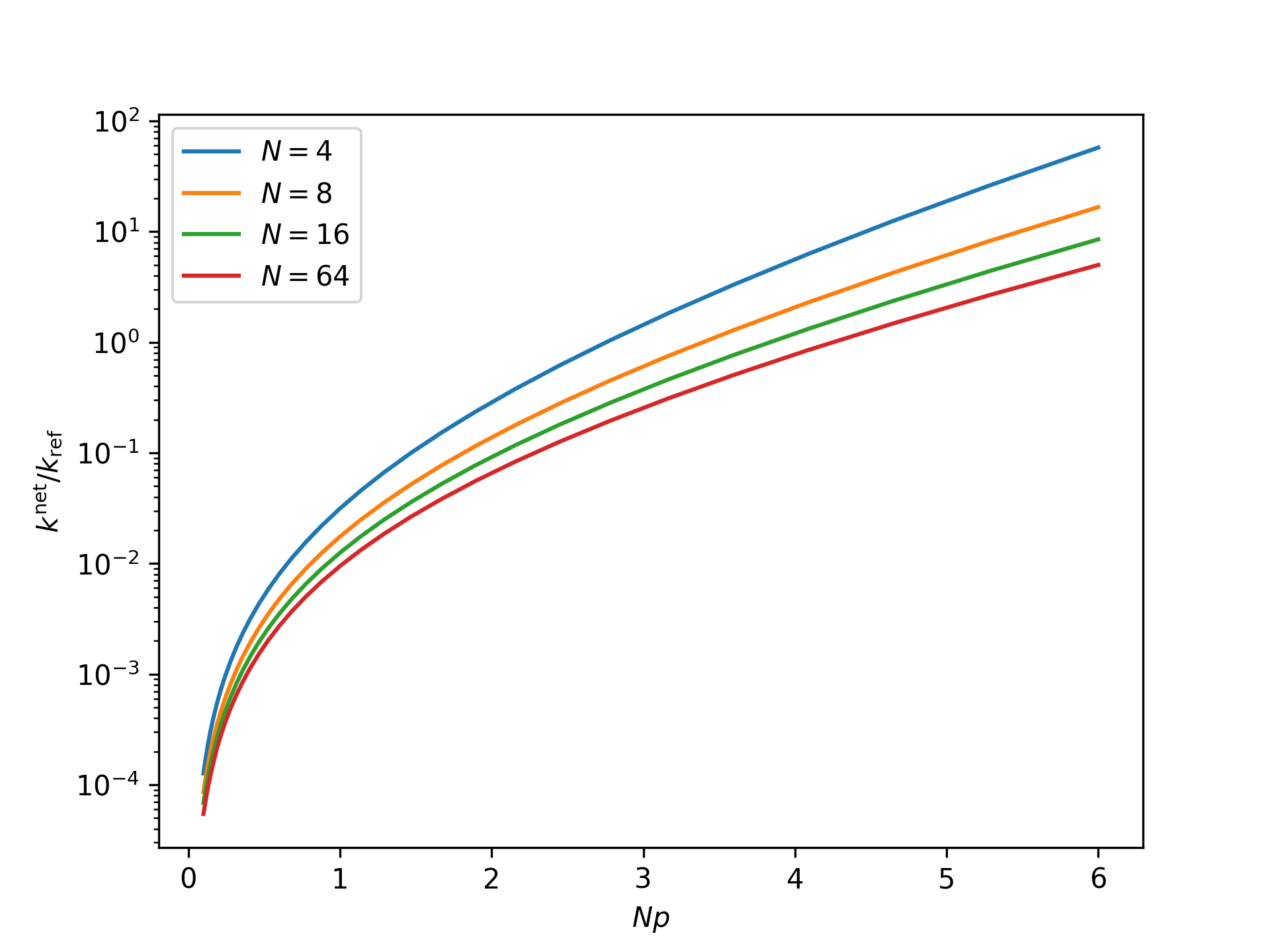
Plot the nondimensional net reaction rate coefficient as a function of the nondimensional end force in the isotensional ensemble for an increasing system size:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import Crack >>> Np = np.logspace(-1, np.log(6)/np.log(10), 33) >>> _ = plt.figure() >>> for N in [4, 8, 16, 64]: ... model = Crack(N=N) ... _ = plt.semilogy( ... Np, model.k_net(Np/N, ensemble='isotensional'), ... label=r'$N=$'+str(N)) >>> _ = plt.xlabel(r'$Np$') >>> _ = plt.ylabel(r'$k^\mathrm{net}/k_\mathrm{ref}$') >>> _ = plt.legend() >>> plt.show()
- k_rev_isotensional(p)[source]
The nondimensional reverse reaction rate coefficient as a function of the nondimensional end force in the isotensional ensemble.
- Parameters:
p (array_like) – The nondimensional end force.
- Returns:
The nondimensional reverse reaction rate.
- Return type:
numpy.ndarray
- v_0_isotensional(p, lambda_)[source]
The nondimensional end separation as a function of the nondimensional end force for the reference system in the isotensional ensemble,
\[v_0(p,\lambda_1,\lambda_2) = -\frac{\partial}{\partial p} \,\beta G_0(p,\lambda_1,\lambda_2) = v_b(p,\lambda_1,\lambda_2).\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (list) – The stretch of the first two intact bonds.
- Returns:
The nondimensional end separation.
- Return type:
numpy.ndarray
Example
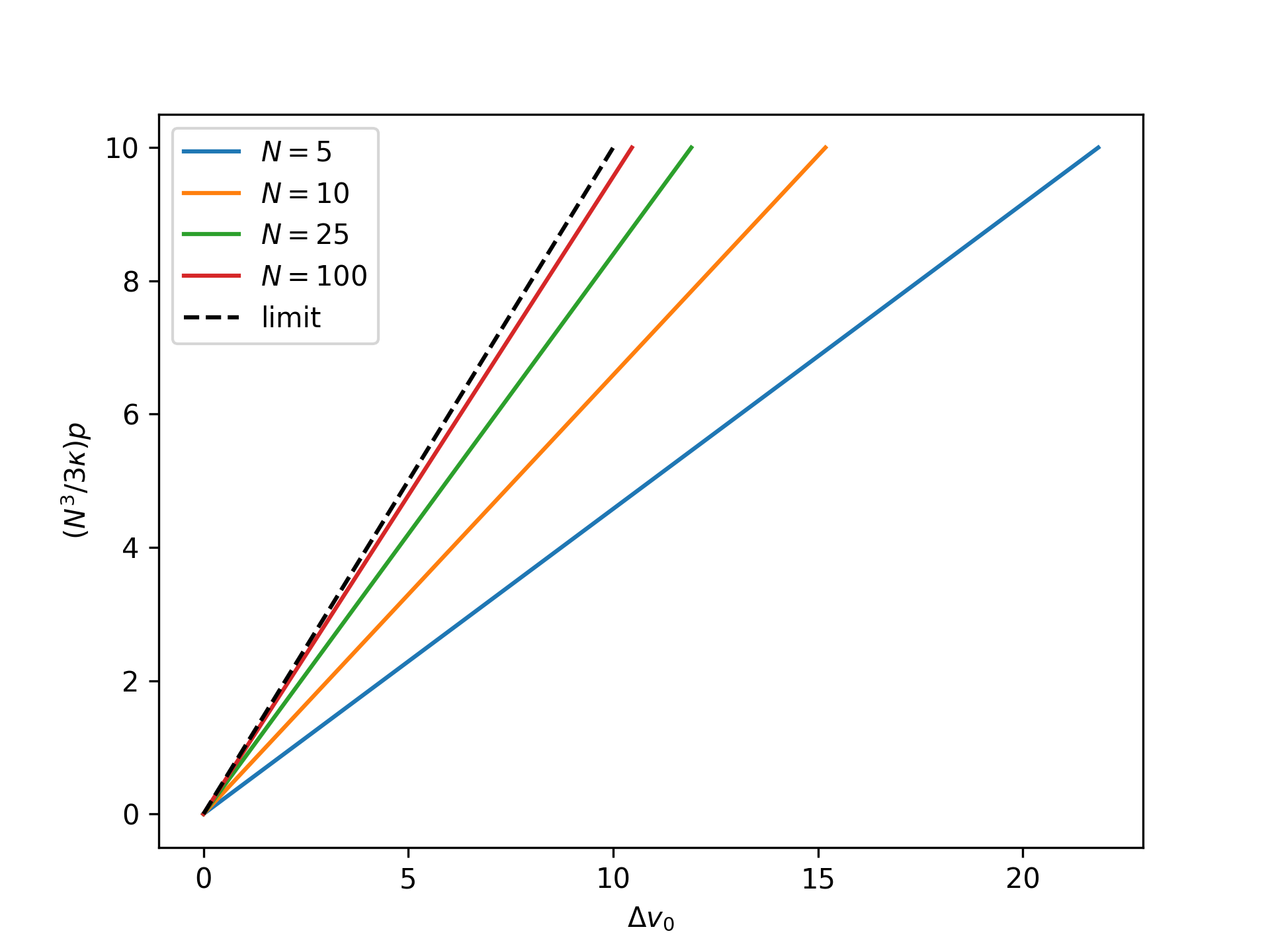
Plot the nondimensional end separation as a function of the rescaled nondimensional end force for the reference system in the isotensional ensemble for an increasing number of broken bonds \(N\) and compare to the thermodynamic limit:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 33) >>> _ = plt.figure() >>> for N in [5, 10, 25, 100]: ... model = CrackIsotensional(N=N) ... p = 3*model.kappa/N**3*rp ... v_0 = model.v_0_isotensional(p, [1, 1]) ... _ = plt.plot(v_0 - 1, rp, label='$N=$'+str(N)) >>> _ = plt.plot(rp, rp, 'k--', label='limit') >>> _ = plt.xlabel(r'$\Delta v_0$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)p$') >>> _ = plt.legend() >>> plt.show()
- v_b_isotensional(p, lambda_)[source]
The nondimensional end separation as a function of the nondimensional end force for the isolated bending system in the isotensional ensemble,
\[v_b(p,\lambda_1,\lambda_2) = -\frac{\partial}{\partial p} \,\beta G_b(p,\lambda_1,\lambda_2).\]In the thermodynamic limit of a large number of broken bonds \(N\), this function has the asymptotic relation
\[v_b(p,\lambda_1,\lambda_2) \sim v_0 + \frac{N^3}{3\kappa}\,p \quad\text{for }N\gg 1.\]- Parameters:
p (array_like) – The nondimensional end force.
lambda (list) – The stretch of the first two intact bonds.
- Returns:
The nondimensional end separation.
- Return type:
numpy.ndarray
- v_isotensional(p, approach='asymptotic', **kwargs)[source]
The nondimensional end separation as a function of the nondimensional end force in the isotensional ensemble,
\[v(p) = -\frac{\partial}{\partial p}\,\beta G(p).\]- Parameters:
p (array_like) – The nondimensional end force.
approach (str, optional, default='asymptotic') – The calculation approach.
**kwargs – Arbitrary keyword arguments. Passed to
v_isotensional_monte_carlo().
- Returns:
The nondimensional end separation.
- Return type:
numpy.ndarray
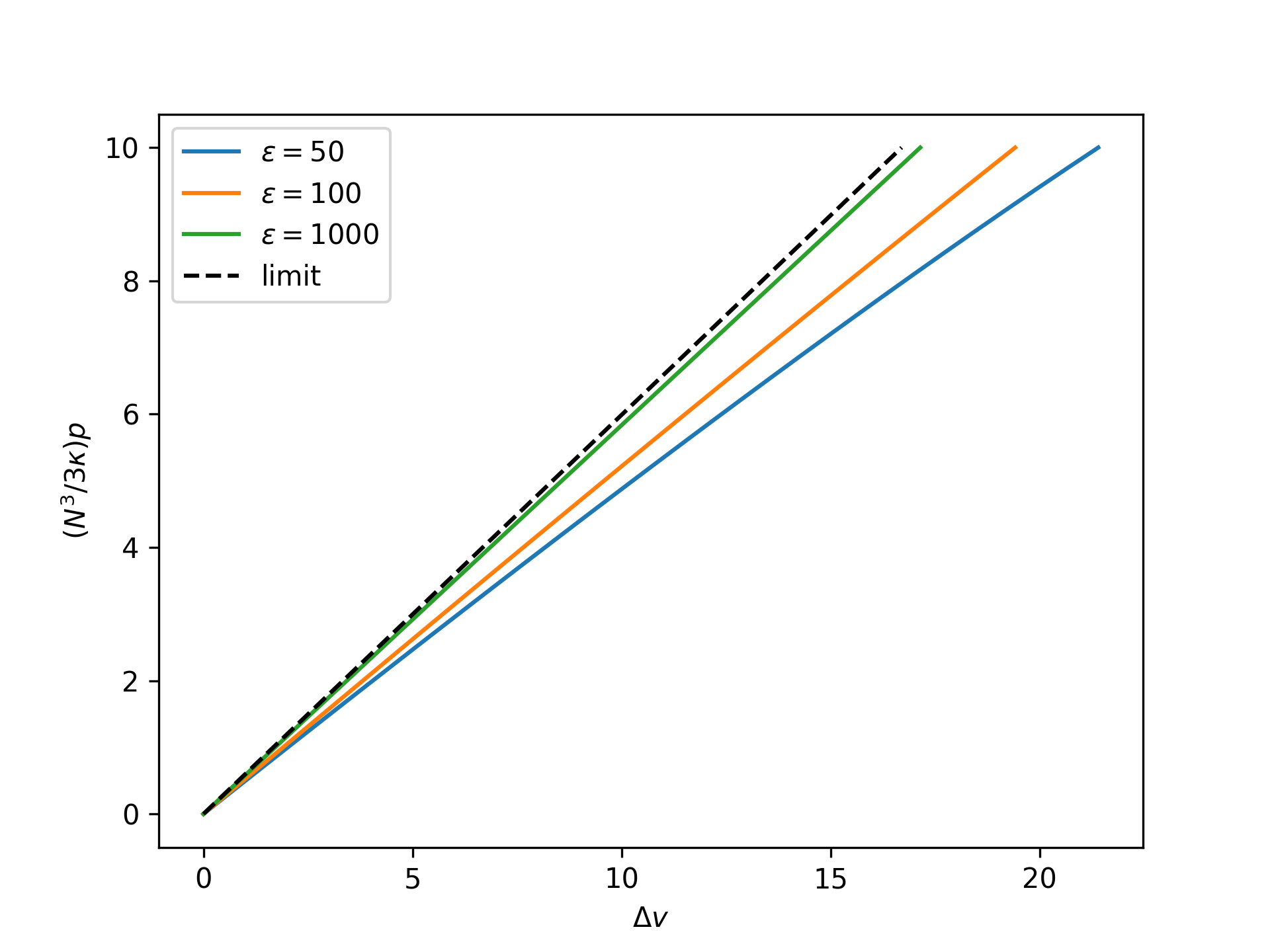
Example
Plot the nondimensional end separation as a function of the rescaled nondimensional end force in the isotensional ensemble for an increasing nondimensional bond energy and compare to the limit given by the reference system:
>>> import numpy as np >>> import matplotlib.pyplot as plt >>> from statmechcrack import CrackIsotensional >>> rp = np.linspace(0, 10, 33) >>> _ = plt.figure() >>> for varepsilon in [50, 100, 1000]: ... model = CrackIsotensional(varepsilon=varepsilon) ... p = 3*model.kappa/model.N**3*rp ... v = model.v_isotensional(p, approach='asymptotic') ... _ = plt.plot(v - 1, rp, ... label=r'$\varepsilon=$'+str(varepsilon)) >>> v_0 = model.v_0_isotensional(p, [1, 1]) >>> _ = plt.plot(v_0 - 1, rp, 'k--', label='limit') >>> _ = plt.xlabel(r'$\Delta v$') >>> _ = plt.ylabel(r'$(N^3/3\kappa)p$') >>> _ = plt.legend() >>> plt.show()